bond_order_processing
Library for processing Mayer bond orders.
The current library allows you to process Mayer bond orders from the CPMD output file
Library version: 1.1.2
Coverage Report
File Stmts Miss Cover Missing main/BondOrderProcessing/bond_order_processing calculations.py 352 9 97% 40, 44, 51, 171, 324, 505, 660, 662–663 calculations_for_atoms_lists.py 166 3 98% 84–87 input_data.py 422 53 87% 88–100, 122–155, 185–189, 225, 234–238, 306, 319–320, 379–383, 430–441, 478–479, 485, 500, 507, 664, 671, 709, 738, 742, 765, 772, 818, 826, 872, 878, 892, 943, 955 TOTAL 941 65 93%
Links
- Documentation page
- Git repository
- More information about CPMD - https://github.com/CPMD-code
You can calculate from Mayer bond orders
- coordinations numbers,
- Qi units,
- Connections between atoms,
- Relation between bond order and length,
- Covalence (The sum of the bond orders of a given atom. This value is close to the valence if atom have only purely covalent bonds)
Example:
>>> from bond_order_processing.input_data import LoadedData >>> from bond_order_processing.calculations import CoordinationNumbers >>> from bond_order_processing.input_data import InputDataFromCPMD >>> >>> path_to_input_file = r'./egzamples_instructions/cpmd_out1.txt' >>> >>> input_data = InputDataFromCPMD() >>> input_data.load_input_data(path_to_input_file, LoadedData.MayerBondOrders) >>> mayer_bond_orders = input_data.return_data(LoadedData.MayerBondOrders) >>> >>> # Calculate percentage of coordination numbers of P >>> coordinations_numbers_stats = CoordinationNumbers.calculate(mayer_bond_orders, 'P', 'O', 'INF', 0.2, 'P-O').calculate_statistics() >>> coordinations_numbers_stats.statistics {4: 100.0}
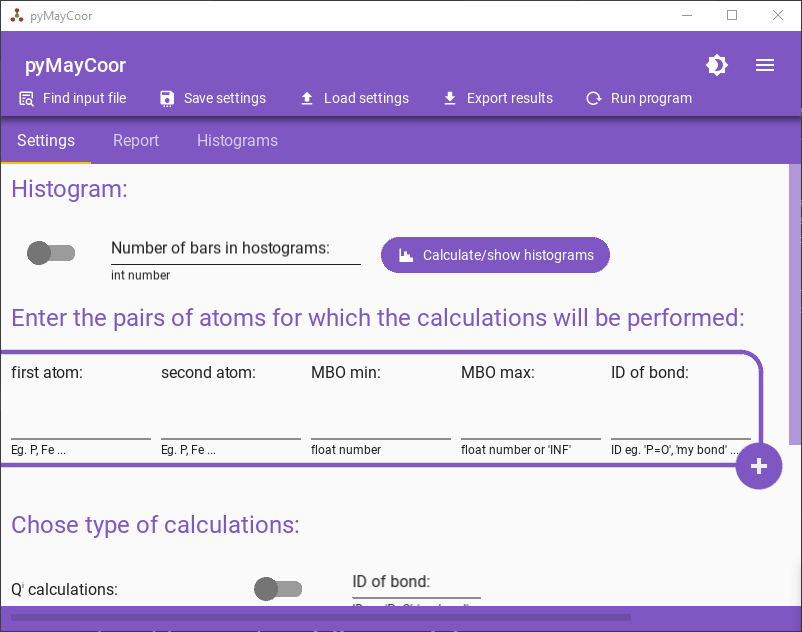
You can also use application using this library.
Check: https://github.com/pawelgoj/pyMayCoor
1"""Library for processing Mayer bond orders. 2 3.. include:: ../README.md 4 5Example: 6 7 >>> from bond_order_processing.input_data import LoadedData 8 >>> from bond_order_processing.calculations import CoordinationNumbers 9 >>> from bond_order_processing.input_data import InputDataFromCPMD 10 >>> 11 >>> path_to_input_file = r'./egzamples_instructions/cpmd_out1.txt' 12 >>> 13 >>> input_data = InputDataFromCPMD() 14 >>> input_data.load_input_data(path_to_input_file, LoadedData.MayerBondOrders) 15 >>> mayer_bond_orders = input_data.return_data(LoadedData.MayerBondOrders) 16 >>> 17 >>> # Calculate percentage of coordination numbers of P 18 >>> coordinations_numbers_stats = CoordinationNumbers.calculate(mayer_bond_orders, 'P', 'O', 'INF', 0.2, 'P-O').calculate_statistics() 19 >>> coordinations_numbers_stats.statistics 20 {4: 100.0} 21 22.. include:: ../app.md 23 24""" 25__docformat__ = "google"
